Google DeepMind researchers Demis Hassabis and John Jumper are jointly sharing half of the 2024 Nobel Prize in Chemistry with David Baker, a biochemist and computational biologist at the University of Washington and Howard Hughes Medical Institute. The Royal Swedish Academy of Sciences selected this year's prize winners for their contributions to protein prediction and design. David Baker was awarded the Nobel Prize for his work on designing entirely new proteins. Hassabis and Jumper were jointly awarded for their work on AlphaFold, which recently predicted the structure of all known proteins.
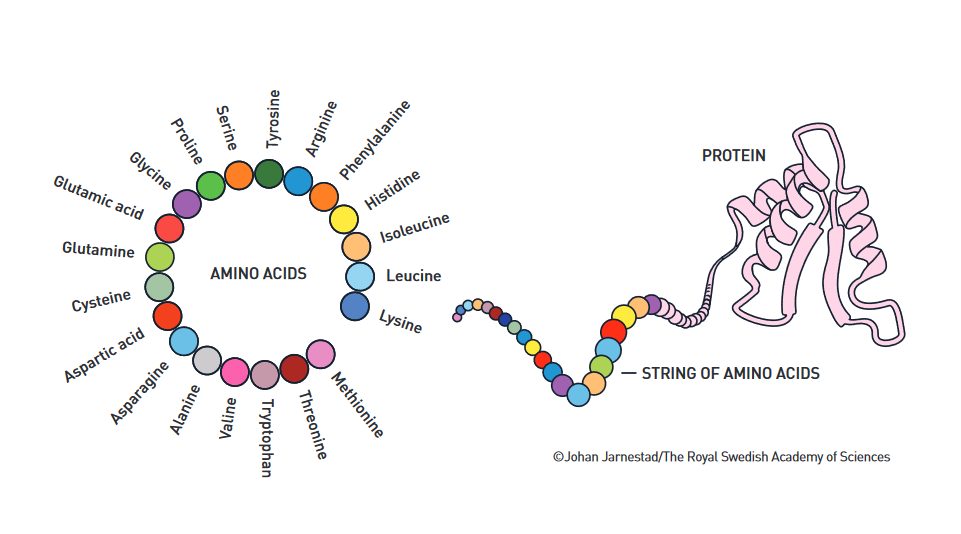
Proteins are often called "the building blocks of life" and with good reason: besides being the main component in most kinds of tissue—including muscle, feathers, hair, and horns—proteins also take on critical roles as enzymes, hormones, signal substances, and antibodies. Due to their importance and ubiquity, researchers have long thought that a better understanding of the inner workings of proteins, and their interactions with other molecules, can unlock beneficial applications in areas including materials science and pharmaceuticals.
David Baker and his research group unlocked a groundbreaking achievement: they designed the world's first de novo protein entirely from scratch. Before this feat, the standard practice was to create de novo proteins using a known structure as a starting point, which limited the range of the results that could be obtained. Baker and his team designed a unique structure without using an existing one as a reference. Then they used computational methods to predict an amino acid chain that would yield the desired structure. Finally, the research group took a gene that encoded the production of the predicted amino acid chain and inserted it into a bacteria that went on to produce the resulting protein.
To confirm their success, the research team determined the produced protein's structure and found that it closely matched the one they had designed at the beginning of the experiment. Thus, Top7, the first de novo protein built entirely from scratch was created. Top7 was the first of many successes attained by Baker's laboratory.
Relatedly, Hassabis, Jumper, and the research team behind AlphaFold wanted to tackle the protein structure prediction problem head-on. To do this, they have developed a series of transformer-based AI models trained on every available database of known protein structures and amino acid sequences. To verify AlphaFold's accuracy, the team registered for the Critical Assessment of Protein Structure Prediction (CASP), a competition where participants were given the amino acid sequences for proteins that had their structure determined but also kept secret. In a competition where no participant had reached over 40% accuracy in their predictions, AlphaFold proved to be almost as accurate as x-ray crystallography, the standard method for determining or confirming a protein's structure.


Comments