The protein folding problem, or at least one of its iterations, consists of reliably determining the 3D structure of a protein from the sequence of its amino acids alone. There is a variety of amino acids serving different purposes, the best known of these molecules being the 22 proteinogenic (protein-creating) amino acids pervasive in the genetic code of all life. For instance, each of the 300,000 known proteins in the human body is encoded by a unique 300-place sequence of these 22 proteinogenic amino acids. Up to this point, the composition of proteins appears to be a relatively simple affair.
From a structural point of view, complexity increases since amino acids attract and repel each other, and these reactions form the distinct folds, loops, and creases that characterize the unique 3D structure of each protein. Moreover, the issue of isolating a protein's structure becomes even more nuanced when one considers that proteins may undergo slight structural changes when carrying out their functions or interacting with other proteins. There is even a category of intrinsically disordered proteins, which, as their name suggests, lack a defined structure in isolation but take on a defined folded shape once they attach to other proteins or molecules as needed to perform their function.
Thus, the experimental determination of a protein's structure is often time-consuming and expensive. Decades of experiments have revealed the structure of nearly 200,000 proteins, and they are all housed in the Protein Data Bank (PDB). The problem is so relevant to science that the CASP (Critical Assessment of Protein Structure Prediction), a community where researchers share their progress in solving the protein folding problem, organizes a biennial challenge where research teams are tasked with predicting the unreleased but experimentally confirmed structure of several proteins.
Armed with the information in the PDB, scientists at Google DeepMind trained AlphaFold so it could predict the undiscovered structure of other proteins based on their amino acid sequence alone. At CASP14 in 2020, AlphaFold's predictions were so accurate that the problem of predicting single protein structures was practically solved. Then, DeepMind released all of AlphaFold's predicted structures to the public domain. In 2021, the database contained over 350,000 entries, including human, yeast, fruit fly, and mouse proteomes. In 2022, the database had been expanded to include predictions for almost every protein known to science. Since its release, AlphaFold's predictions have helped catalyze research into new malaria vaccines, the discovery of new cancer drugs, and the development of plastic-eating enzymes.
Now, DeepMind and Isomorphic Labs have announced the publication of a progress update on the latest version of AlphaFold: it can now predict the structure of nearly every molecule in the PDB, often with atomic accuracy. In addition to proteins, these molecules include ligands, nucleic acids, and molecules that include post-translational modifications (PTMs). In this iteration, AlphaMind tackles specific protein structure prediction problems, such as antibody binding or protein-ligand complexes. The current standard for the latter involves using docking methods that require a rigid reference protein and a suggested binding position for the ligand. AlphaFold outperformed docking method predictions without requiring a reference or an approximate location for the ligand to bind to.
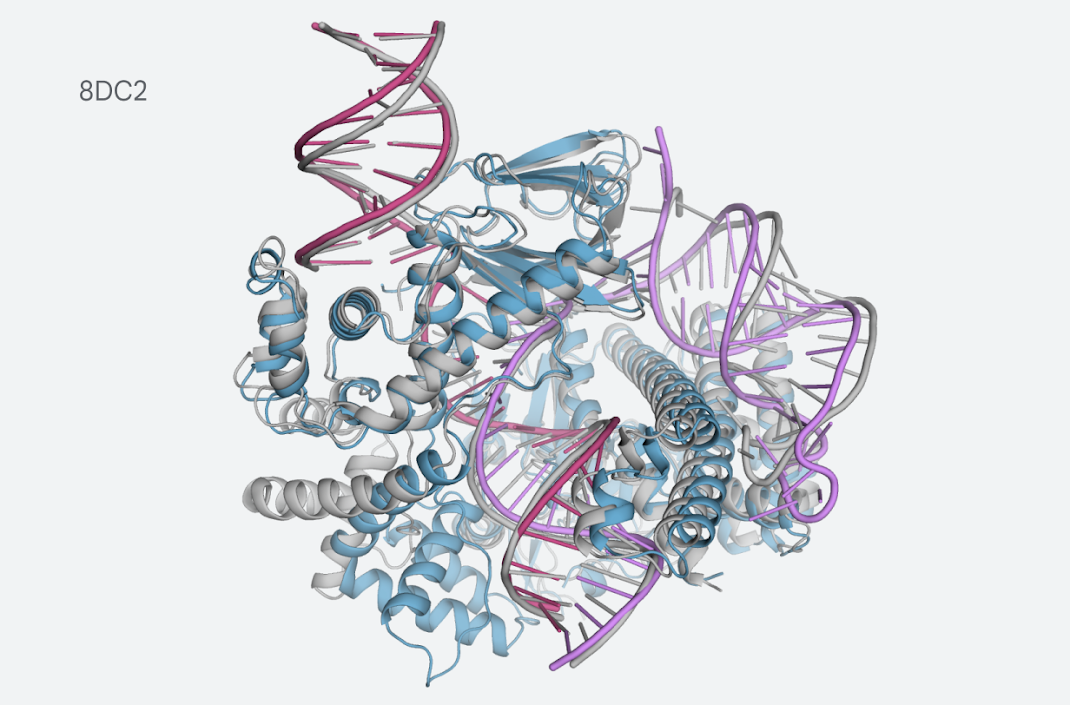
The progress update includes a thorough report on the model inputs and outputs, as well as the evaluation process. Essentially, the model was trained on data available on the PDB up to the cutoff date of September 30, 2021. The evaluation set incorporates data available past the cutoff date, including a subset determined to be low homology to the training set (in other words, the data most different from the one contained in the training set). Performance results focus on the low homology subset. One of the most notable examples is the prediction for the CasLambda bound to crRNA and DNA. CasLambda belongs to the CRISPR family and shares the gene-editing capabilities of the CRISPR-Cas9 system but is smaller, which has led scientists to think it may be a more efficient gene-editing particle.
If anything, the progress update shows that AlphaFold has come a long way since its first release and will probably continue to accelerate the advancement of scientific exploration in many fields. It is also important to acknowledge the critical voices reminding us that predictions are different from experimentally verified data and should be handled with an additional measure of caution. Indeed, AlphaFold is by no means a replacement for experimentally confirmed results. But even when the problem is obtaining more experimentally confirmed data, the model can positively impact research by, for instance, helping scientists optimize resource allocation to focus on researching molecules on which AlphaFold has a low confidence score. This only goes to show that the model is nothing short of extraordinary.




Comments